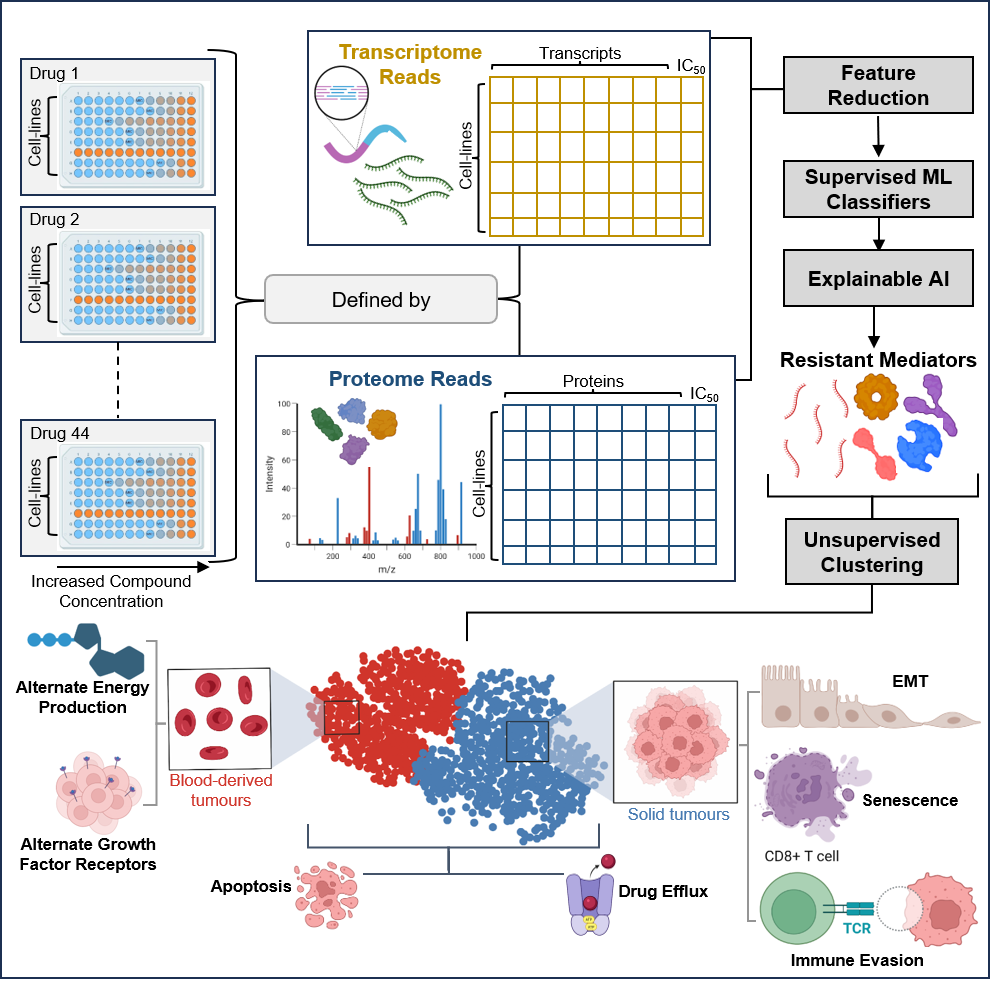
In this study, we conducted a comprehensive, high-throughput analysis using supervised machine learning classifiers applied to transcriptomic and proteomic data from pan cancer cell lines classified as resistant or sensitive to FDA-approved targeted drugs. Among the evaluated drugs, Axitinib demonstrated particularly high predictive accuracy.
To uncover the molecular determinants of resistance, we employed explainable AI algorithm: LIME, identifying key molecular players associated with resistance. Further, to understand various mechanisms of resistance, unsupervised clustering was performed. Enrichment analysis further revealed the biological pathways in which these resistance mediators are overrepresented, and pathway cross-talk was examined to elucidate the perturbed biological processes.
Repository Organisation
📂 Axitinib_Codes_2025_may/
├── 📂 1_DataProcessing/
│ ├── 📂 1_GDSC_Drugs_Preprocessing/
│ └── 📄 GDSC_Processing_and_EDA.ipynb
│ ├── 📂 2_Data_Representation_Transcriptomics_TPM/
│ └── 📄 Data_Representation_Transcriptomics_TPM.ipynb
│ ├── 📂 3_Data_Representation_Protein_Intensity/
│ └── 📄 Data_Representation_Protein_Intensity.ipynb
│ └── 📂 4_Targeted_Drugs_FDA_Approved/
│ └── 📄 Targeted_Drugs_Processing.ipynb
│
├── 📂 2_AutomatedFeatureEnggAndML/
│ ├── 📂 Proteomics/
│ │ ├── 📂 Prot_BatchA/
│ │ ├── 📄 Result_Post-processing.ipynb
│ │ └── 📄 automationbatchcode_protbatchA.py
│ │ └── 📂 Prot_BatchB/
│ │ ├── 📄 Result_Post-processing.ipynb
│ │ └── 📄 automationbatchcode_protbatchB.py
│ └── 📂 Transcriptomics/
│ ├── 📂 Trans_BatchA/
│ │ ├── 📄 Result_Processing.ipynb
│ │ └── 📄 automationbatchcode_transBatchA.py
│ └── 📂 Trans_BatchB/
│ ├── 📄 Result_Processing.ipynb
│ └── 📄 automationbatchcode_transbatchb.py
│
├── 📂 3_BestPerformingDrugProcessing/
│ └── 📄 ComparisonofAllAutomatedvsSelectedDrugs_MLScores.ipynb
│
├── 📂 4_ExplainableAI_LIME/
│ ├── 📄 Transcriptomics_Lime.ipynb
│ └── 📄 Proteomics_Lime.ipynb
│
├── 📂 5_Correlation/
│ ├── 📄 Correlation_Resistancecell_Lines_T.ipynb
│ └── 📄 Correlation_Resistancecell_Lines_P.ipynb
│
└── 📂 6_Clustering_and_StatisticalAnalysis/
├── 📄 Clustering.ipynb
├── 📄 Same_Clusters.ipynb
├── 📄 Variance_Analysis.ipynb
└── 📄 Mean_Analysis.ipynb
==================================================
TOTAL CODE FILES: 21
==================================================
