In this study, we developed a deep learning model to classify medical images obtained from various modalities (X-ray, Ultrasound, Histopathological and Dermoscopic images). This project aims to detect medical image anomalies with good accuracy and aid in early diagnosis, thus contributing to the advancements in medical informatics.
We have built the model on backbone architectures of foundation model based encoder, supplemented by nonlinear transformations of the image features and robust feature extraction using multi-scale attention module. The performance across all the medical datasets were evaluated and meaning biological patterns were inferred with explainable AI which proved the real world application of the model.
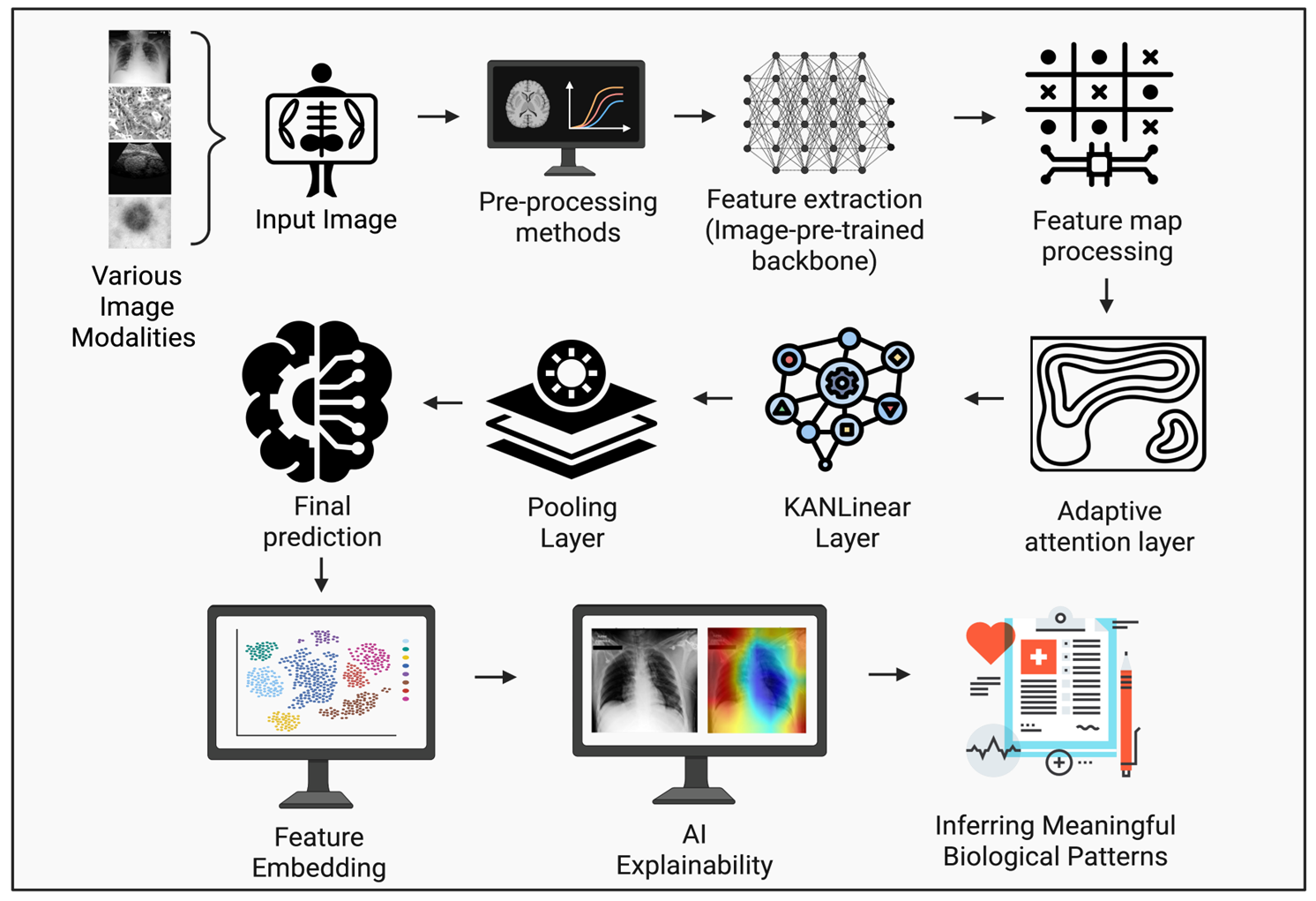
Repository Organisation
📂 Image_Classification/
├── 📂 1_Model_Architecture/
│ ├── 📂 1_Global_pooling/
│ │ └── 📄 Global_pooling.py
│ ├── 📂 2_Attention_Module/
│ │ └── 📄 XSAM_Attention.py
│ ├── 📂 3_KANLinear_Transformation/
│ │ └── 📄 KANLinear.py
│ └── 📂 4_Model_Compilation/
│ └── 📄 volo_model.py
│
├── 📂 2_Foundation_Models/
│ └── 📂 Architecture_Backbones/
│ ├── 📄 conv_former.py
│ ├── 📄 eva_model.py
│ ├── 📄 mvit_model.py
│ ├── 📄 nextvit_model.py
│ └── 📄 pvt_model.py
│
├── 📂 3_Ablation_Study/
│ ├── 📂 1_Model_No_Attention/
│ │ └── 📄 ablation_without_attention.py
│ ├── 📂 2_Model_No_KANLinear_Transformation/
│ │ └── 📄 ablation_without_KAN.py
│ └── 📂 3_Model_with_KAN_Attention/
│ └── 📄 model_attention_KAN.py
│
├── 📂 4_Data_Pre_Processing/
│ └── 📄 pre-processing.ipynb
│
├── 📂 5_Training_and_Evaluation/
│ ├── 📂 1_Training_and_Validation/
│ │ ├── 📄 train_validation.py
│ │ └── 📄 loss_function.py
│ └── 📂 2_Evaluation_Metric_Visualization/
│ └── 📄 evaluation_metrices.ipynb
│
├── 📂 6_Explainable_AI_and_Feature_Embedding/
│ ├── 📂 1_Explainable_AI/
│ │ ├── 📄 GradCAM.py
│ │ ├── 📄 Guided_Saliency_Map.py
│ │ └── 📄 Vanilla_Gradient_Saliency_Map.py
│ └── 📂 2_Feature_Embedding/
│ └── 📄 tSNE.ipynb
│
├── 📄 Dataset_details.txt
├── 📄 LICENSE
├── 📄 README.md
└── 📄 requirements.txt
==================================================
TOTAL CODE FILES: 17
==================================================
