In this comprehensive study, we conducted a thorough evaluation of large language models for medical question answering, specifically focusing on Wilson's Disease. Our research employed advanced retrieval methods including BM25, TF-IDF, and PubMedBERT embeddings, combined with ensemble approaches to enhance information retrieval accuracy.
We evaluated seven state-of-the-art language models across multiple metrics (ROUGE-L, BLEU, METEOR) in both zero-shot and few-shot settings, generating a high-quality synthetic QA dataset with 217 question-answer pairs. The pipeline demonstrates exceptional performance with the best ensemble method achieving 69.1% recall@7 and minimal hallucination rates.
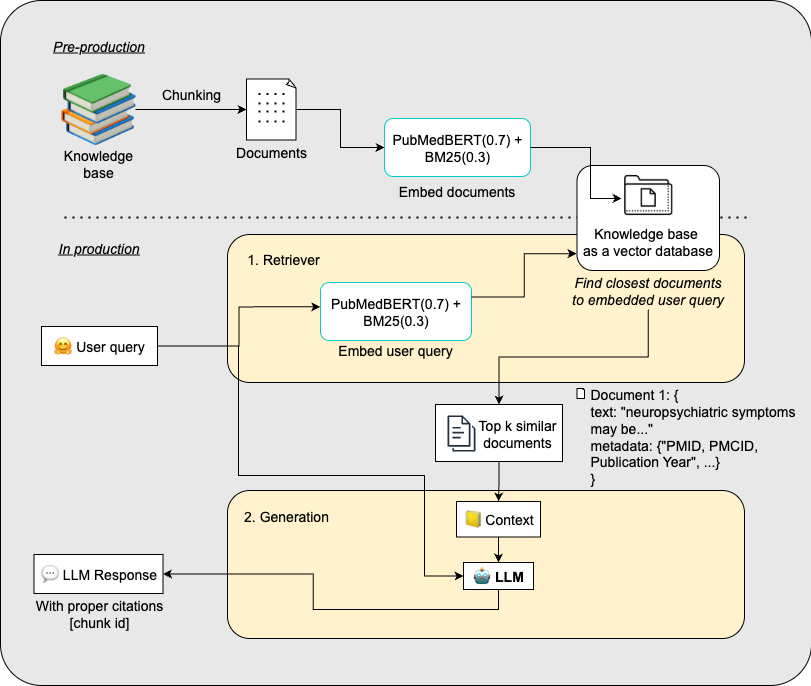
RAG System Architecture
Repository Organisation
📂 Wilson_Disease_Medical_QA_2025/
├── 📂 Data&DatasetProcessing/
│ ├── 📄 Data_Collection.ipynb
│ ├── 📄 Data_Preprocessing.ipynb
│ ├── 📄 Data_Chunking.ipynb
│ ├── 📄 Data_Indexing_and_Retrieval_Evaluation.py
│ ├── 📄 Dataset_Generation_and_Evaluation_WD_QA.ipynb
│ ├── 📄 Data_Serialisation.py
│ ├── 📄 README.md
│ ├── 📊 evaluation_summary_statistics.csv
│ ├── 📊 correlation_heatmap.png
│ ├── 📊 score_boxplot.png
│ ├── 📊 score_distributions.png
│ ├── 📊 preprocessed_papers.csv
│ ├── 📊 serialized_retriever.pkl
│ ├── 📊 articles_with_fulltext.csv
│ ├── 📊 preprocessed_papers_with_chunks.csv
│ ├── 📊 qa_evaluation_results.csv
│ ├── 📊 qa_pairs_processed.csv
│ ├── 📊 qa_evaluation_results_filtered.csv
│ ├── 📊 all.csv
│ ├── 📊 score_summary_statistics.csv
│ ├── 📊 score_distribution_counts.csv
│ └── 📊 Retriever_Evaluation_Summary.csv
│
├── 📂 Generation&Benchmarking/
│ ├── 📂 ZeroShot/
│ │ ├── 📄 gpt-4o-mini-2024-07-18.py
│ │ ├── 📄 claude-3-7-sonnet-20250219.py
│ │ ├── 📄 Biomistral-Instructv0.2-7B-DARE.py
│ │ ├── 📄 Qwen2.5-7B-Instruct.py
│ │ ├── 📄 drllama-7b.py
│ │ ├── 📄 deepseek-llm-7b-chat.py
│ │ ├── 📄 Llama-3.2-3B-Instruct-MedicalQA.py
│ │ ├── 📄 gemma-3-4b-it.py
│ │ ├── 📊 gpt-4o-mini-2024-07-18.csv
│ │ ├── 📊 claude-3-7-sonnet-20250219.csv
│ │ ├── 📊 Biomistral-Instructv0.2-7B-DARE.csv
│ │ ├── 📊 Qwen2.5-7B-Instruct.csv
│ │ ├── 📊 drllama-7b.csv
│ │ ├── 📊 deepseek-llm-7b-chat.csv
│ │ └── 📊 Llama-3.2-3B-Instruct-MedicalQA.csv
│ ├── 📂 FewShot/
│ │ ├── 📄 gpt-4o-mini-2024-07-18.py
│ │ ├── 📄 claude-3-7-sonnet-20250219.py
│ │ ├── 📄 Biomistral-Instructv0.2-7B-DARE.py
│ │ ├── 📄 Qwen2.5-7B-Instruct.py
│ │ ├── 📄 drllama-7b.py
│ │ ├── 📄 deepseek-llm-7b-chat.py
│ │ ├── 📄 Llama-3.2-3B-Instruct-MedicalQA.py
│ │ ├── 📄 gemma-3-4b-it.py
│ │ ├── 📊 gpt-4o-mini-2024-07-18.csv
│ │ ├── 📊 claude-3-7-sonnet-20250219.csv
│ │ ├── 📊 Biomistral-Instructv0.2-7B-DARE.csv
│ │ ├── 📊 Qwen2.5-7B-Instruct.csv
│ │ ├── 📊 drllama-7b.csv
│ │ ├── 📊 deepseek-llm-7b-chat.csv
│ │ └── 📊 Llama-3.2-3B-Instruct-MedicalQA.csv
│ ├── 📂 evaluation_results/
│ │ ├── 📊 metrics_summary_table.txt
│ │ ├── 📊 comparison_max.csv
│ │ ├── 📊 comparison_max.txt
│ │ ├── 📊 comparison_min.csv
│ │ ├── 📊 comparison_min.txt
│ │ ├── 📊 comparison_median.csv
│ │ ├── 📊 comparison_median.csv
│ │ ├── 📊 comparison_std.csv
│ │ ├── 📊 comparison_mean.csv
│ │ ├── 📊 comparison_std.txt
│ │ ├── 📊 comparison_mean.txt
│ │ ├── 📊 shot_improvement.png
│ │ ├── 📊 meteor_median_comparison.png
│ │ ├── 📊 meteor_mean_comparison.png
│ │ ├── 📊 bleu_median_comparison.png
│ │ ├── 📊 bleu_mean_comparison.png
│ │ ├── 📊 rouge_l_median_comparison.png
│ │ ├── 📊 rouge_l_mean_comparison.png
│ │ └── 📊 metrics_data.json
│ ├── 📄 evaluate.py
│ ├── 📄 Prompts.md
│ ├── 📄 README.md
│ └── 📊 serialized_retriever.pkl
│
└── 📂 Misc_SourceFiles/
├── 📄 DataSourceFile.ipynb
├── 📄 simple_timeline_plot.py
├── 📊 wilson_disease_timeline.png
└── 📊 PubMed_Timeline_Results_by_Year.csv
==================================================
TOTAL CODE FILES: 29
==================================================
